Example 2: StarDist (Detection) + Stracking (Tracker)
This example shows a combination of StarDist Detection and Stracking (Tracker)
Load trained mode
from stardist.models import StarDist2D
# prints a list of available models
StarDist2D.from_pretrained()
# creates a pretrained model
model = StarDist2D.from_pretrained('2D_versatile_fluo')
There are 4 registered models for 'StarDist2D':
Name Alias(es)
──── ─────────
'2D_versatile_fluo' 'Versatile (fluorescent nuclei)'
'2D_versatile_he' 'Versatile (H&E nuclei)'
'2D_paper_dsb2018' 'DSB 2018 (from StarDist 2D paper)'
'2D_demo' None
Found model '2D_versatile_fluo' for 'StarDist2D'.
Loading network weights from 'weights_best.h5'.
Loading thresholds from 'thresholds.json'.
Using default values: prob_thresh=0.479071, nms_thresh=0.3.
StarDist: Prediction and detection
from csbdeep.utils import normalize
import matplotlib.pyplot as plt
from tifffile import imread
import numpy as np
from stracking.detectors import SSegDetector
folder=""
filename="P31-crop2.tif"
img= imread(folder+filename)
labels=np.zeros(img.shape);
img=normalize(img)
for i in range(img.shape[0]):
labels[i,:,:],details = model.predict_instances(img[i,:,:])
sdetector = SSegDetector(is_mask=False)
particles = sdetector.run(labels)
Create an empty napari viewer
%gui qt
import napari
from napari.utils import nbscreenshot
viewer = napari.Viewer(axis_labels='tyx')
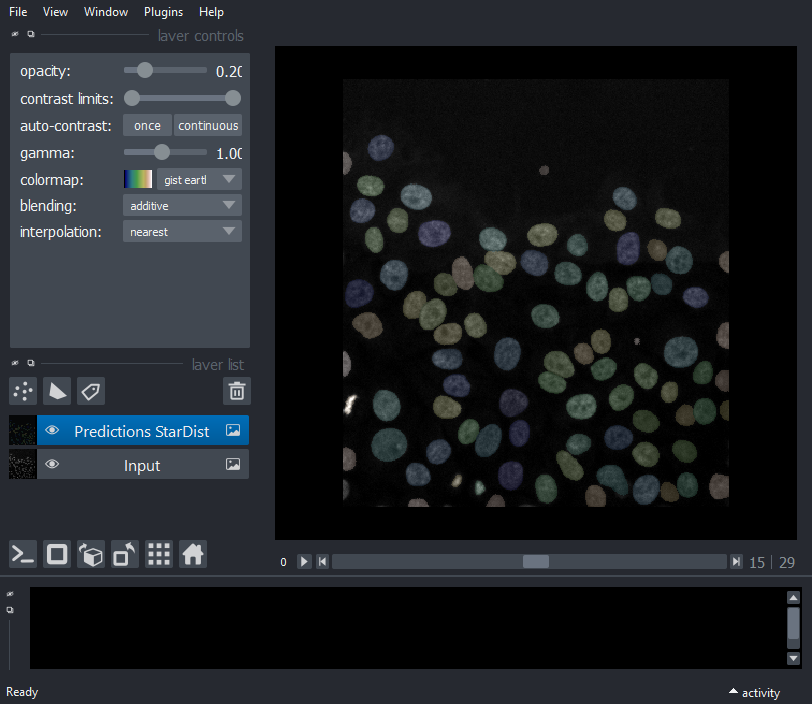
Display Input and StarDist Prediction
viewer.add_image(img, name='Input', multiscale=False,
contrast_limits=[0, 3], colormap='gray',blending='additive');
viewer.add_image(labels, name='Predictions StarDist', multiscale=False,
colormap='gist_earth',blending='additive',opacity=0.2);
nbscreenshot(viewer)
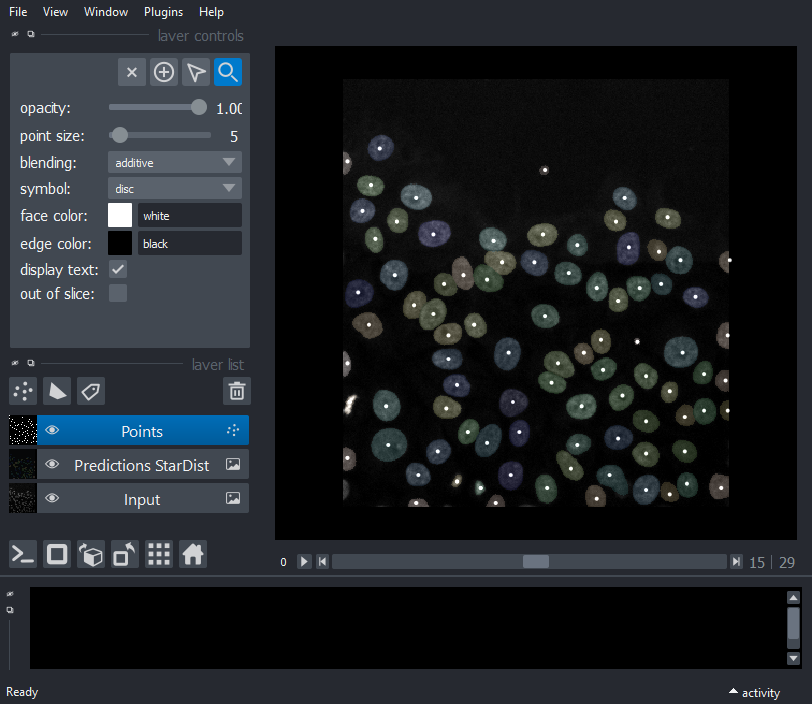
Display spots from StarDist
viewer.add_points(particles.data, size=5, blending='additive')
nbscreenshot(viewer)
Linker
Shortest path tracking with euclidean cost
from stracking.linkers import SPLinker, EuclideanCost
euclidean_cost = EuclideanCost(max_cost=100);
my_tracker = SPLinker(cost=euclidean_cost, gap=1);
tracks = my_tracker.run(particles);
detections shape= (2652, 3)
num frames= 30
cost= 60244.0
self.cost.max_cost= 100
cost= 256.0
self.cost.max_cost= 100
cost= 16.0
self.cost.max_cost= 100
cost= 11236.0
self.cost.max_cost= 100
extract track...
dim in track to path= 2
add predecessor...
add predecessor...
add predecessor...
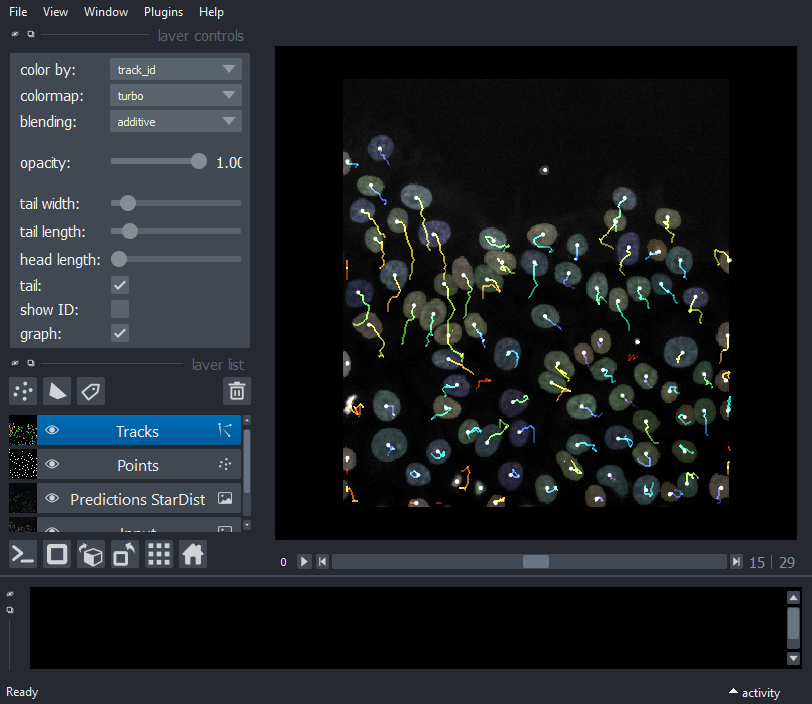
Display tracks
viewer.add_tracks(tracks.data, name='Tracks')
nbscreenshot(viewer)