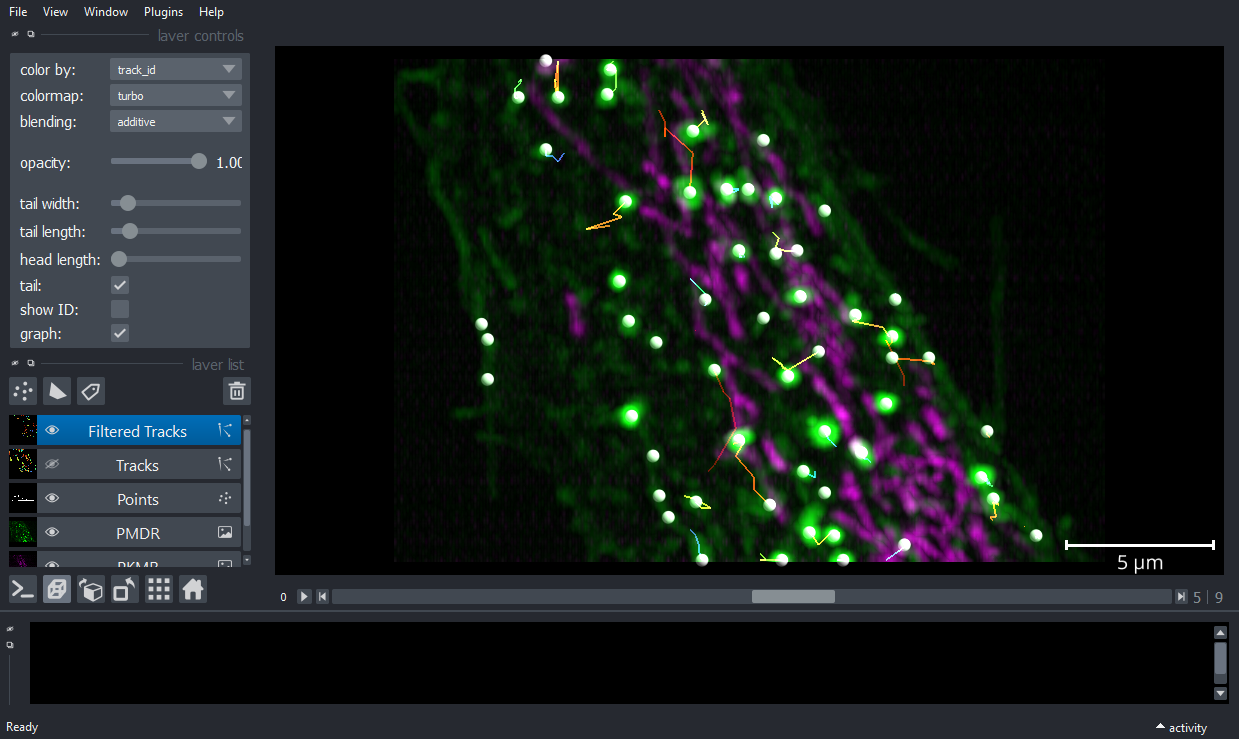
Example 1: Stracking workflow
This example shows how to detect particles in 3D+t image using the LoG detector
Load data
from stracking import data
from tifffile import imread
folder=""
filenamePKMR="PKMR_10timepoints_crop.tif"
filenamePMDR="PMDR_10timepoints_crop.tif"
PKMRimg= imread(folder+filenamePKMR)
PMDRimg= imread(folder+filenamePMDR)
Create an empty napari viewer
%gui qt
import napari
from napari.utils import nbscreenshot
viewer = napari.Viewer(axis_labels='tzyx')
Display volumetric timeseries
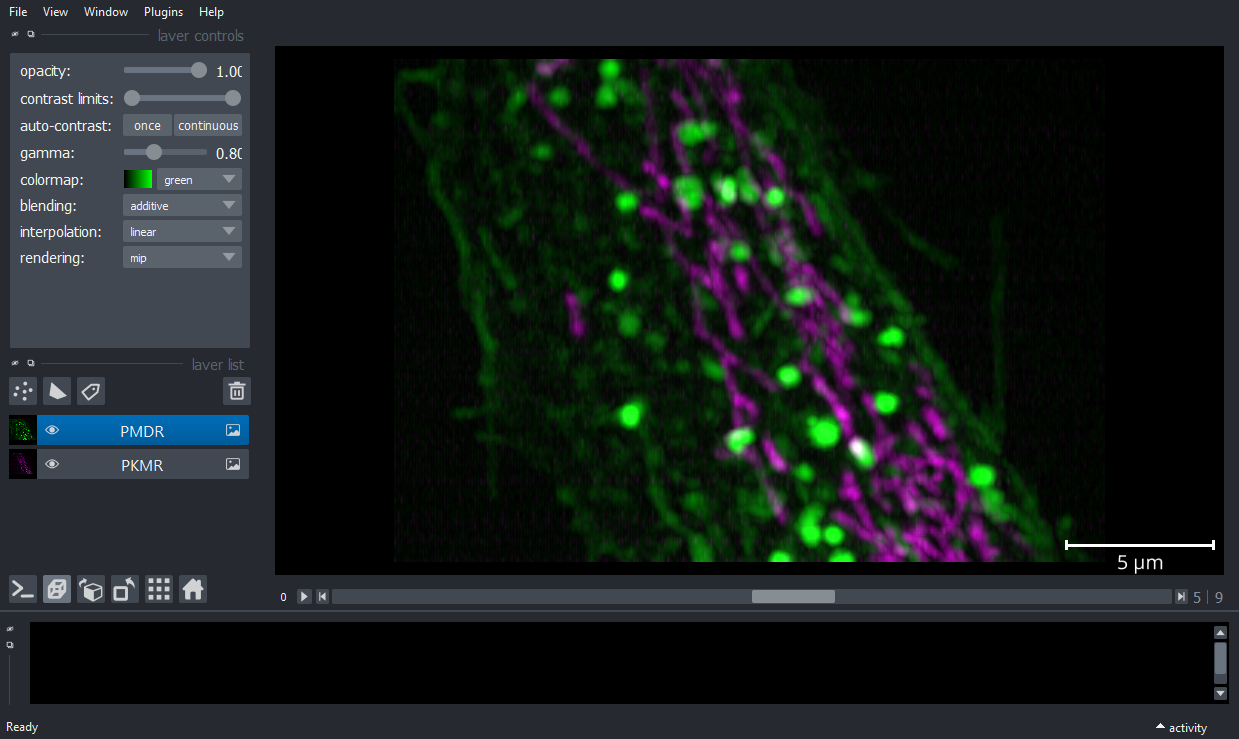
viewer.add_image(PKMRimg, name='PKMR', multiscale=False, scale=[4.3,0.316,0.104,0.104],
contrast_limits=[10, 600], colormap='magenta',blending='additive');
viewer.add_image(PMDRimg, name='PMDR', multiscale=False, scale=[4.3,0.316,0.104,0.104],
contrast_limits=[10, 1_000], colormap='green',blending='additive',gamma=0.8);
viewer.dims.ndisplay = 3
viewer.scale_bar.visible='true'
viewer.scale_bar.unit='um'
nbscreenshot(viewer)
LoG 3D+t detection
from stracking.detectors import LoGDetector
detector = LoGDetector(min_sigma=3, max_sigma=5, num_sigma=3, threshold=0.001)
particles = detector.run(PMDRimg)
Display spots
viewer.add_points(particles.data, size=5, shading='spherical',scale=[4.3,0.316,0.104,0.104],blending='additive')
nbscreenshot(viewer)

Spots properties
from stracking.properties import IntensityProperty
property_calc = IntensityProperty(radius=2.5)
property_calc.run(particles,PKMRimg)
y=particles.properties['mean_intensity']
particlesch1=particles
property_calc = IntensityProperty(radius=2.5)
property_calc.run(particles,PMDRimg)
x=particles.properties['mean_intensity']
Spots statistics
import matplotlib.pyplot as plt
import numpy as np
from matplotlib import colors
from matplotlib.ticker import PercentFormatter
import seaborn as sns
plt.style.use('_mpl-gallery')
fig, axs = plt.subplots(1, 2, figsize=(15, 4), sharey=False)
axs[0].set_title("Mean Intensity [a.u.]",fontsize=14)
axs[0].set_ylabel("Mitochondria Intensity",fontsize=14)
axs[0].set_xlabel("Endosomes Intensity",fontsize=14)
axs[0].scatter(x,y,alpha=0.5,color='r');
axs[0].tick_params(axis='both', labelsize=14)
sns.set_style("whitegrid")
axs[1] = sns.swarmplot(y=x,alpha=0.9,size=3)
axs[1].set_ylabel("Endosomes Intensity",fontsize=14)
axs[1].set_xlabel("PMDR",fontsize=14)
axs[1].set_title("Endosomes Intensity [a.u.]",fontsize=14)
axs[1].tick_params(axis='both',labelsize=14)

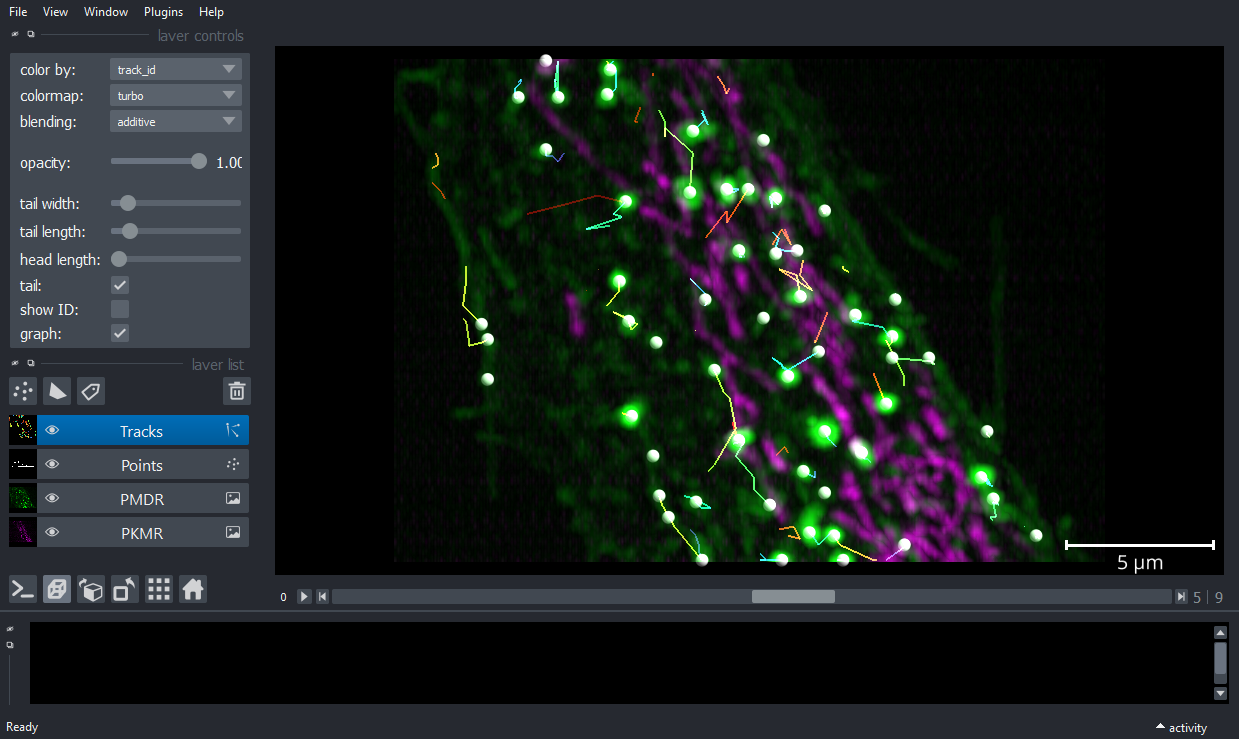
Tracker
Shortest path tracking with euclidean cost
from stracking.linkers import SPLinker, EuclideanCost
euclidean_cost = EuclideanCost(max_cost=225)
my_tracker = SPLinker(cost=euclidean_cost, gap=1)
tracks = my_tracker.run(particles)
detections shape= (564, 4)
num frames= 10
cost= 1.0
self.cost.max_cost= 225
cost= 2819.0
self.cost.max_cost= 225
cost= 5433.0
self.cost.max_cost= 225
cost= 8438.0
self.cost.max_cost= 225
extract track...
dim in track to path= 3
add predecessor...
add predecessor...
Display tracks
viewer.add_tracks(tracks.data, name='Tracks',scale=[4.3,0.316,0.104,0.104])
nbscreenshot(viewer)
Features - tracking
from stracking.features import (LengthFeature, DistanceFeature,
DisplacementFeature)
tracks.scale=[1,0.316,0.104,0.104];
# Length feature
feature_calc = LengthFeature()
tracks = feature_calc.run(tracks)
# Distance feature
feature_calc = DistanceFeature()
tracks = feature_calc.run(tracks)
# Displacement feature
feature_calc = DisplacementFeature()
tracks = feature_calc.run(tracks)
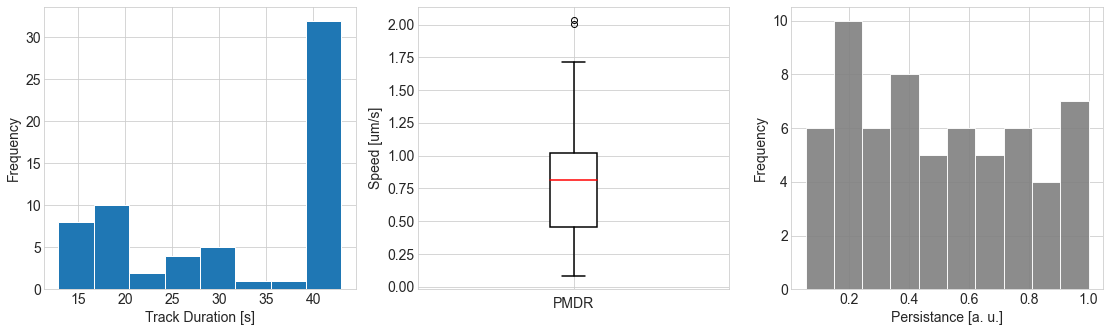
Display tracks properties
displacement=tracks.features['displacement'];
distance=tracks.features['distance'];
length=tracks.features['length'];
displacementMat = np.array([displacement[i] for i in range(len(displacement))])
distanceMat = np.array([distance[i] for i in range(len(distance))])
lengthMat = np.array([length[i] for i in range(len(length))])
speed=distanceMat/(4.3*lengthMat)
fig, axs = plt.subplots(1, 3, figsize=(15, 4), sharey=False)
axs[0].hist(4.3*lengthMat, bins=8, edgecolor="white")
axs[0].set_xlabel("Track Duration [s]",fontsize=14)
axs[0].set_ylabel("Frequency",fontsize=14)
axs[0].tick_params(axis='both', labelsize=14)
axs[1].boxplot([speed],patch_artist=True,
medianprops={"color": "red", "linewidth": 1.5},
boxprops={"facecolor": "white", "edgecolor": "black",
"linewidth": 1.5},
whiskerprops={"color": "black", "linewidth": 1.5},
capprops={"color": "black", "linewidth": 1.5},labels=[''])
axs[1].set_ylabel("Speed [um/s]",fontsize=14)
axs[1].set_xlabel("PMDR",fontsize=14)
axs[1].tick_params(axis='both', labelsize=14)
axs[2].hist(displacementMat/distanceMat, bins=10, edgecolor="white",facecolor='gray',alpha=0.9)
axs[2].set_xlabel("Persistance [a. u.]",fontsize=14)
axs[2].set_ylabel("Frequency",fontsize=14)
axs[2].tick_params(axis='both', labelsize=14)
Features tracking
from stracking.filters import FeatureFilter
f_filter = FeatureFilter(feature_name='length', min_val=10, max_val=100)
filtered_tracks = f_filter.run(tracks)
viewer.add_tracks(filtered_tracks.data,features=filtered_tracks.features,name='Filtered Tracks',scale=[4.3,0.316,0.104,0.104])
viewer.layers['Tracks'].visible=False
nbscreenshot(viewer)